You can now upload files
How would you expect the 2 trees to render? Currently, only the second one is shown because both have the same coordinates.
You can show that by using a mask, where we hide 2 cells at level 1 on each piece
mask = pack_binary_string("0 0011 00000000 000 0 1100 00000000 000")
[...]
vtkgrp.create_dataset(
"Mask",
data=mask,
dtype='u1',
shape=mask.shape,
maxshape=(None,)
)
(1 value for each cell + byte padding)
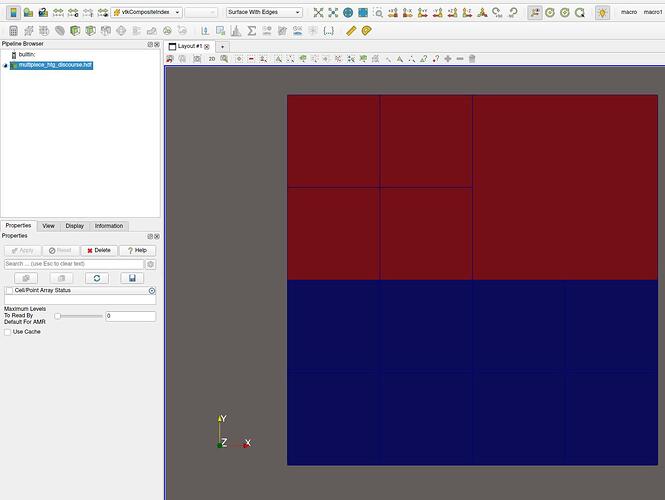
On ParaView with composite index as coloring:
Yes, I was hoping for some result like the image. The actual code I am using does not necessarily share the same descriptors for the tree on all processes and so for now I have been using MPIIO trickery to write a single “piece” file. I did try something similar previously with multiblock datasets and the mask field(s) but it seems that VTK/Paraview filters treated these as discontinuous.
Is it safe to say if using masking with the multipiece HTG datasets will treat CellData in the composed tree as continuous? Admittedly I haven’t tried this out yet.
What do you mean by “continuous”? Which ParaView filters do you expect to work with your multi-piece dataset?
If we’re talking contours or ghost cell generation, the computation may not cross the multi-block boundary, but I assume it would for multi-piece HyperTreeGrids
Mostly with regards to the most common ones e.g. contours/isosurfaces. Previously my group was mostly writing the tree as an unstructured grid which had a connectivity issue w.r.t. hanging nodes. It has been hopeful so far to see that HTG solves this issue but I am still hoping to get a writer working that doesn’t rely massively on MPI/MPIIO coordination to put everything into a single file.
Note that for the vtkHDFWriter class we provide, for MPI parallel writing, we don’t rely on MPIIO, but each rank will create a file on its own with its data. Then, when all ranks are done writing, rank 0 will create a “main” file that glues datasets together using HDF5 “Virtual Datasets”, basically soft-linking datasets from each piece/rank to make a single file, without hard copies of the data. This is an approach you may want to try as well.