Hi,
Indeed I haven’t been able to notice a difference when strip.SetJoinContiguousSegments() is set to True.
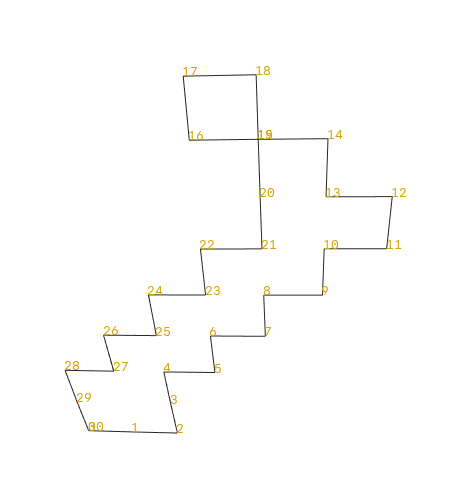
How to sort anti-clockwise points from a polydata ?
You will see that points 16, 17, 18 are not ordered correctly if you expect an anti-clockwise sort.
from vedo import *
! wget -q -nc https://thredds-su.ipsl.fr/thredds/fileServer/ipsl_thredds/brocksce/tmp/poly_02.vtk
g = load('poly_02.vtk')
line = Line(g.points())
labels = line.labels2d(c="yellow4", scale=0.7)
show(line, labels, axes=0, interactive=True, zoom=1, roll=90, azimuth=-90, elevation=0)