Hi there,
my VTK lessons are failing me, so asking this here.
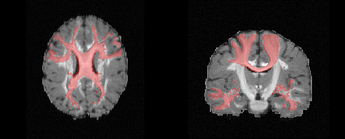
I have an anatomical image of a brain, and a segmentation of a region of interest. Both saved as NIfTI files.
I would like to be able to compute and visualize the intersection of an arbitrary plane (e.g. axial, coronal, sagittal at any given slice) on the contour of the segmentation. When the normal of the clipping plane is not a given one, there seems to be no intersection, whereas I expect there should be one.
Maybe that has to do with the fact that my volume is not centered at the origin. I see that my plane and the contour are far apart when visualizing, but I do set the origin of my cutting plane to the center of my image data. Furthermore, I guess that if that were an issue, it should also affect to the plane normal value that works.
Any guess what step I’m missing to make it work for any arbitrary plane/normal?
I’m using Python and VTK 9.1.0. Attached some pictures illustrating the volume data and the segmentation, the result of the clip with a normal plane that actually works, and the script. The NIfTI files of interest can be found here:
https://drive.google.com/drive/folders/12RjvUInGlU9vEttZjoyCRU-MBqQX_i0Q?usp=sharing
I’m using nibabel to load the NIfTI files.
The script can be called as:
python clip_plane_test.py T1.nii.gz CC.nii.gz
Thanks for the time.
clip_plane_test.py (5.0 KB)