Hi, I’m working on creating a segmentation pipeline and am having issues with creating a closed .stl file from my segmentations. The code block is below and here is a picture of the .stl that is created. As you can see it looks good from the top view.
However, the bottom view isn’t closed and the outer rings aren’t interpolating to the inner one.
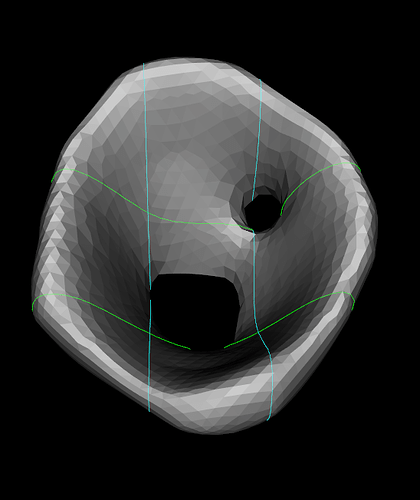
The desired bottom should be something like this (taken in 3D Slicer)

Any help is appreciated!
Code:
def process_nifti_files_to_stl(self):
nifti_path = os.path.join('combined_output', 'combined_volume.nii.gz')
if not os.path.exists(nifti_path):
QMessageBox.warning(self, "File Not Found", "No combined NIfTI file found. Please create it first.")
return
sitk_image = sitk.ReadImage(nifti_path)
image_array = sitk.GetArrayFromImage(sitk_image)
input_image = image_from_array(image_array)
interpolated_image = morphological_contour_interpolation(
input_image=input_image,
label=1,
axis=-1,
no_heuristic_alignment=False,
no_use_distance_transform = False,
use_custom_slice_positions=False,
no_use_extrapolation=False,
use_ball_structuring_element=False
)
interpolated_array = interpolated_image.data
vtk_image = vtk.vtkImageData()
depth, height, width = interpolated_array.shape
vtk_image.SetDimensions(width, height, depth)
vtk_image.SetSpacing(sitk_image.GetSpacing())
flat_image_array = interpolated_array.flatten()
vtk_data_array = numpy_to_vtk(num_array=flat_image_array, deep=True, array_type=vtk.VTK_FLOAT)
vtk_image.GetPointData().SetScalars(vtk_data_array)
print(vtk_image.GetNumberOfPoints())
print(vtk_image.GetPointData().GetScalars().GetRange())
gaussianRadius = 0.75
gaussianStandardDeviation = 1.5
gaussian = vtk.vtkImageGaussianSmooth()
gaussian.SetInputData(vtk_image)
gaussian.SetStandardDeviations(
gaussianStandardDeviation,
gaussianStandardDeviation,
gaussianStandardDeviation
)
gaussian.SetRadiusFactors(gaussianRadius, gaussianRadius, gaussianRadius)
gaussian.Update()
print(gaussian.GetOutput().GetNumberOfPoints())
print(gaussian.GetOutput().GetPointData().GetScalars().GetRange())
isoValue = gaussian.GetOutput().GetPointData().GetScalars().GetRange()[1] / 2 # Adjust as needed
use_flying_edges = True
if use_flying_edges:
print("Using vtkFlyingEdges3D for isosurface extraction.")
iso_surface = vtk.vtkFlyingEdges3D()
else:
print("Using vtkMarchingCubes for isosurface extraction.")
iso_surface = vtk.vtkMarchingCubes()
iso_surface.SetInputConnection(gaussian.GetOutputPort())
iso_surface.ComputeScalarsOff()
iso_surface.ComputeGradientsOff()
iso_surface.ComputeNormalsOff()
iso_surface.SetValue(0, isoValue)
iso_surface.Update()
smoothingIterations = 20
passBand = 0.001
featureAngle = 60.0
smoother = vtk.vtkWindowedSincPolyDataFilter()
smoother.SetInputConnection(iso_surface.GetOutputPort())
smoother.SetNumberOfIterations(smoothingIterations)
smoother.BoundarySmoothingOff()
smoother.FeatureEdgeSmoothingOff()
smoother.SetFeatureAngle(featureAngle)
smoother.SetPassBand(passBand)
smoother.NonManifoldSmoothingOn()
smoother.NormalizeCoordinatesOn()
smoother.Update()
final_poly_data = smoother.GetOutput()
stl_writer = vtk.vtkSTLWriter()
stl_output_path = os.path.join('combined_output', 'combined_volume_smoothed.stl')
stl_writer.SetFileName(stl_output_path)
stl_writer.SetInputData(final_poly_data)
stl_writer.Write()