Hi guys,
I import dicom data Using vtkDICOMImageReader(). The 3-plane result is upside-down, as follows:
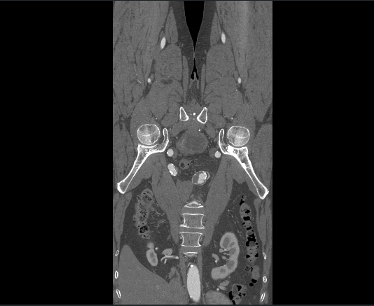
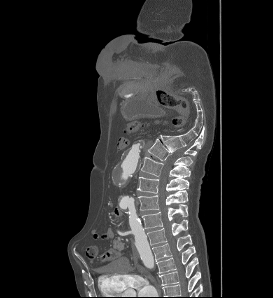
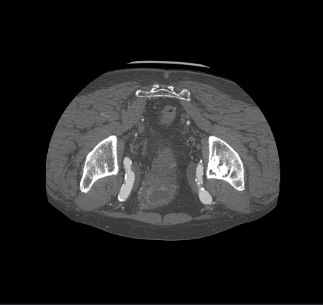
But if I convert the DICOM files to .mha or .nii.gz, and using vtk to visualize them, there is no upside-down problem.
I have read many documents about this problem.The most related I found is the below one:
https://www.nitrc.org/plugins/mwiki/index.php/dcm2nii:MainPage#Spatial_Coordinates
According to this ,DICOM images are saved to disk in the order we read English: with the first line at the top of the screen and subsequent lines descending.
It seems the reason cause the problem.
But here is my question: Is there a standard inside way to deal with this using vtk? Or we must use transform to get the volume direction right ?
Ps: I know extend dicom support using vtk-dicom, but I’m using python. As far as I kown, the package don’t support python.