OK. This is a closed surface mesh, which should be no problem to convert to binary labelmap. You can do it using VTK, using vtkPolyDataToImageStencil class as shown here.
If you find it hard to figure out the necessary coordinate system transforms then you can use the converter in 3D Slicer. Using the GUI: load the image data and model (vtk) files, create segmentation for the image (right-click on image -> Segment this), import the model into the segmentation (drag-and-drop the model to the segmentation node), then export the segmentation to binary labelmap (right-click on segmentation -> Export to binary labelmap):
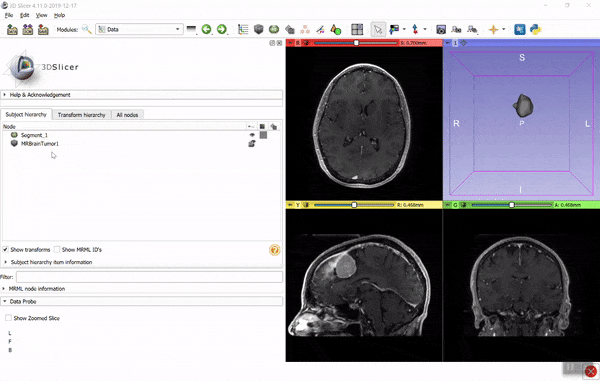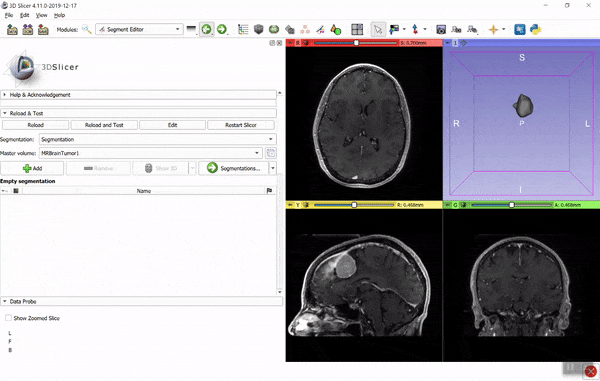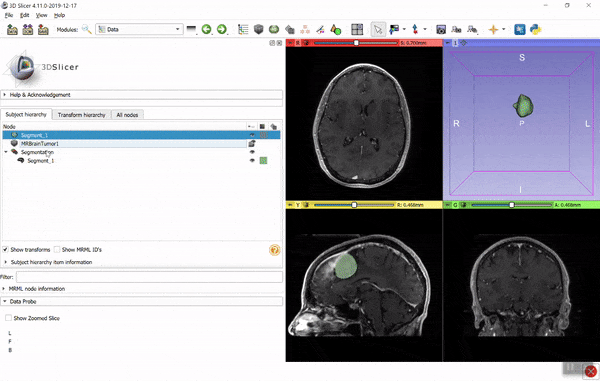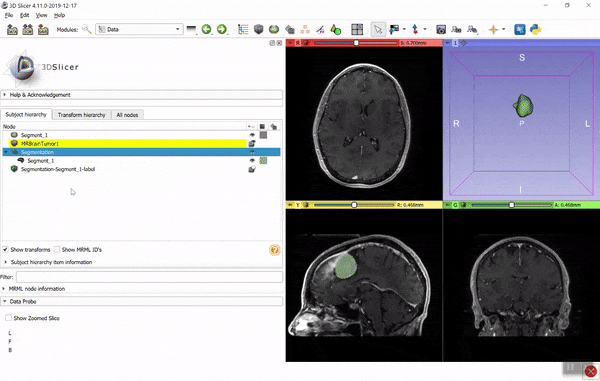
You can save the binary labelmap as a nrrd or nifti file or access it directly as a numpy array. You can automate all the above steps using a few lines of Python script (see examples here). If you have any questions related to 3D Slicer, post it to Slicer forum.
If you do the segmentation in MITK Workbench then probably you can directly save it as a nrrd or nifti file (that you can load as a numpy array, without any conversion).
I’ve also noticed that the segmentation is very low quality, due to the highly anisotropic input image (image spacing is about 5-10x larger along one of the axis). You may want to resample your input volume to be isotropic before segmenting it (you can do this in 3D Slicer using Crop volume module, by enabling “Isotropic spacing” option) to be able to create smoother, more detailed segmentation.