Hello everyone and thank you for your answers,
I’m a new user of VTK. And excuse my english.
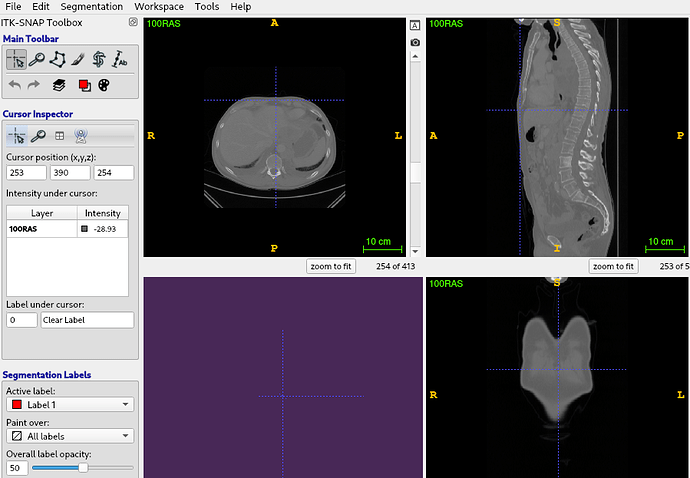
I have a niftii image volume with many landmarks. I want to reslice this image with an affine transformation matrix, at this point i use this program and that’s work well.
import vtk
import numpy as np
import random
import pandas as pd
import scipy.stats
import numpy.linalg as npl
from datetime import datetime
from functions import *
random.seed(datetime.now())
def samples_pdf(df):
samples = []
for dfhead in df.columns:
samples.append(scipy.stats.gaussian_kde(df_9var[str(dfhead)].to_numpy()))
return samples
def generate_mat_9var(s):
getsamp = lambda samples, i: samples[i].resample(1).T[:,0][0]
MTrans = np.array([
[getsamp(s,0), getsamp(s,1), getsamp(s,2), 0.],
[getsamp(s,3), getsamp(s,4), getsamp(s,5), 0.],
[getsamp(s,6), getsamp(s,7), getsamp(s,8), 0.],
[0., 0., 0., 1.]
])
return MTrans
""" Return mm point (world) to voxel point """
def mmtovox(pointmm, target_affine):
return (nib.affines.apply_affine(npl.inv(target_affine), pointmm))
""" Return voxel point to mm point (world) """
def voxtomm(pointvox, target_affine):
return nib.affines.apply_affine(target_affine, pointvox)
df_9var = pd.read_csv('9var.csv', sep=',', error_bad_lines=False)
samples = samples_pdf(df_9var)
print("Open image")
file = "../../DATA/100RAS.nii.gz"
reader = vtk.vtkNIFTIImageReader()
reader.SetFileName(file)
reader.Update()
Output = reader.GetOutput()
print("Make transform")
bounds = np.array(Output.GetBounds())
transform = vtk.vtkTransform()
center = np.zeros(3)
transformCenter = np.zeros(3)
for i in range( len(center) ):
center[i] = 0.5 * (bounds[i*2]+bounds[i*2+1])
Mat = generate_mat_9var(samples)
transform.SetMatrix(Mat.flatten())
print("Reslice Image")
reslice = vtk.vtkImageReslice()
reslice.SetInputData( Output )
reslice.AutoCropOutputOn();
reslice.SetResliceTransform( transform )
reslice.SetBackgroundLevel( Output.GetPointData().GetScalars().GetRange()[0] )
reslice.Update()
print("Save Image")
savevol(reslice)
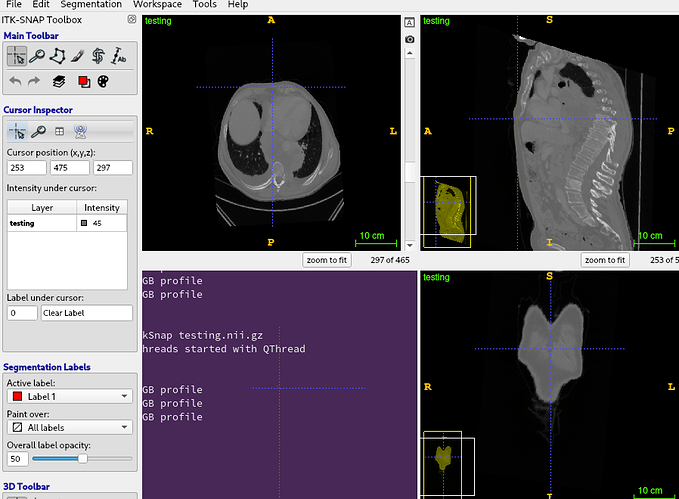
But when I want to obtain the new point coordinates, that’s not work.
I’ve tried some calcul like
MyNewPoint = np.Dot( MyActualPoint, Mat) + origin
Anyone can help me pleaz ?