In the blessed month of Ramadan, I was Alhamdu lillah able to make something in Julia which works:
Julia code
using HDF5
using StaticArrays
fType = Float64
idType = Int64
connectivities = [“Vertices”, “Lines”, “Polygons”, “Strips”]
Positions = SVector{3, Float64}[
[0.0, 0.0, 0.5],
[0.0, 0.0, -0.5],
[0.5, 0.0, 3.061617e-17],
[-0.25, 0.4330127, 3.061617e-17],
[-0.25, -0.4330127, 3.061617e-17]
]
Normals = SVector{3, Float64}[
[0.0, 0.0, 1.0],
[0.0, 0.0, -1.0],
[1.0, 0.0, 6.123234e-17],
[-0.5, 0.8660254, 6.123234e-17],
[-0.5, -0.8660254, 6.123234e-17]
]
Warping = SVector{3, Float64}[
[0.0, 0.0, 0.0],
[0.0, -0.0, 0.0],
[0.0, -0.5, 0.0],
[0.4330127, 0.25, -0.0],
[-0.4330127, 0.25, 0.0]
]
function generate_geometry_structure(root)
# Write version of VTKHDF format as an attribute
HDF5.attrs(root)[“Version”] = Int32.([2, 3])
# Write type of dataset ("PolyData") as an ASCII string to a "Type" attribute.
# This is a bit tricky because VTK/ParaView don't support UTF-8 strings here, which is
# the default in HDF5.jl.
let s = "PolyData"
dtype = HDF5.datatype(s)
HDF5.API.h5t_set_cset(dtype.id, HDF5.API.H5T_CSET_ASCII)
dspace = HDF5.dataspace(s)
attr = HDF5.create_attribute(root, "Type", dtype, dspace)
HDF5.write_attribute(attr, dtype, s)
end
NumberOfPoints = HDF5.create_dataset(root, "NumberOfPoints" , idType , ((0,),(-1,)), chunk=(100,))
Points = HDF5.create_dataset(root, "Points" , fType , ((3,0), (3,-1)), chunk=(3,100))
for connect in connectivities
group = HDF5.create_group(root, connect)
NumberOfConnectivityIds = HDF5.create_dataset(group, "NumberOfConnectivityIds", idType, ((0,),(-1,)), chunk=(100,) )
NumberOfCells = HDF5.create_dataset(group, "NumberOfCells", idType, ((0,),(-1,)), chunk=(100,) )
Offsets = HDF5.create_dataset(group, "Offsets", idType, ((0,),(-1,)), chunk=(100,) )
Connectivity = HDF5.create_dataset(group, "Connectivity", idType, ((0,),(-1,)), chunk=(100,) )
end
pData = HDF5.create_group(root, "PointData")
Warping = HDF5.create_dataset(pData, "Warping", fType, ((3,0),(3,-1)), chunk=(3,100))
Normals = HDF5.create_dataset(pData, "Normals", fType, ((3,0),(3,-1)), chunk=(3,100))
return nothing
end
function generate_step_structure(root)
steps = HDF5.create_group(root, “Steps”)
NSteps, _ = HDF5.create_attribute(steps, "NSteps", Int32)
Values = HDF5.create_dataset(steps, "Values", fType , ((0,),(-1,)), chunk=(100,))
singleDSs = ["PartOffsets", "NumberOfParts", "PointOffsets"]
for name in singleDSs
HDF5.create_dataset(steps, name, idType, ((0,),(-1,)), chunk=(100,))
end
nTopoDSs = ["CellOffsets", "ConnectivityIdOffsets"]
for name in nTopoDSs
HDF5.create_dataset(steps, name, idType, ((4,0),(4, -1)), chunk=(4, 100))
end
pData = HDF5.create_group(steps, "PointDataOffsets")
Warping = HDF5.create_dataset(pData, "Warping", idType, ((0,),(-1,)), chunk=(100,))
Normals = HDF5.create_dataset(pData, "Normals", idType, ((0,),(-1,)), chunk=(100,))
end
function append_data(root, newStep, Positions)
steps = root[“Steps”]
# To update attributes, this is the best way I've found so far
old_NSteps = HDF5.read_attribute(steps, "NSteps")
steps_attr_dict = attributes(steps)
NSteps = steps_attr_dict["NSteps"]
write_attribute(NSteps,HDF5.datatype(idType), old_NSteps + 1)
HDF5.set_extent_dims(steps["Values"], (length(steps["Values"]) + 1,))
steps["Values"][end] = newStep
PointsStartIndex = size(root["Points"])[2] + 1
PositionLength = length(Positions)
HDF5.set_extent_dims(root["Points"], (length(first(Positions)), size(root["Points"])[2] + PositionLength))
root["Points"][:, PointsStartIndex:(PointsStartIndex+PositionLength-1)] = stack(Positions) * rand()
HDF5.set_extent_dims(steps["PointOffsets"], (length(steps["PointOffsets"]) + 1,))
steps["PointOffsets"][end] = PointsStartIndex - 1
HDF5.set_extent_dims(root["NumberOfPoints"], (length(root["NumberOfPoints"]) + 1,))
root["NumberOfPoints"][end] = PositionLength
NumberOfPartsStartIndex = length(steps["NumberOfParts"]) + 1
HDF5.set_extent_dims(steps["NumberOfParts"], (length(steps["NumberOfParts"]) + 1,))
steps["NumberOfParts"][NumberOfPartsStartIndex] = 1
PartOffsetsStartIndex = length(steps["PartOffsets"]) + 1
PartOffsetsLength = length(steps["PartOffsets"]) + 1
HDF5.set_extent_dims(steps["PartOffsets"], (PartOffsetsLength,))
steps["PartOffsets"][PartOffsetsStartIndex] = PartOffsetsLength - 1
CellOffsetsStartIndex = size(steps["CellOffsets"])[2] + 1
HDF5.set_extent_dims(steps["CellOffsets"], (4, CellOffsetsStartIndex))
steps["CellOffsets"][:, CellOffsetsStartIndex] = zeros(4)
ConnectivityIdOffsetsStartIndex = size(steps["ConnectivityIdOffsets"])[2] + 1
HDF5.set_extent_dims(steps["ConnectivityIdOffsets"], (4, ConnectivityIdOffsetsStartIndex))
steps["ConnectivityIdOffsets"][:, ConnectivityIdOffsetsStartIndex] = zeros(4)
NumberOfPartsStartIndex = length(steps["NumberOfParts"]) + 1
HDF5.set_extent_dims(steps["NumberOfParts"], (length(steps["NumberOfParts"]) + 1,))
steps["NumberOfParts"][NumberOfPartsStartIndex] = 1
for point_data_name in keys(steps["PointDataOffsets"])
HDF5.set_extent_dims(steps["PointDataOffsets"][point_data_name], (length(steps["PointDataOffsets"][point_data_name]) + 1,))
steps["PointDataOffsets"][point_data_name][end] = PointsStartIndex - 1
end
HDF5.set_extent_dims(root["PointData"]["Normals"], (length(first(Normals)), size(root["PointData"]["Normals"])[2] + length(Normals)))
root["PointData"]["Normals"][:, PointsStartIndex:(PointsStartIndex+PositionLength-1)] = stack(Normals) * rand()
HDF5.set_extent_dims(root["PointData"]["Warping"], (length(first(Warping)), size(root["PointData"]["Warping"])[2] + length(Warping)))
root["PointData"]["Warping"][:, PointsStartIndex:(PointsStartIndex+PositionLength-1)] = stack(Warping) * rand()
for connect in connectivities
for dataset in ["NumberOfCells", "NumberOfConnectivityIds", "Offsets", "Connectivity"]
HDF5.set_extent_dims(root[connect][dataset], (length(root[connect][dataset]) + 1,))
root[connect][dataset][end] = idType(0)
end
end
end
function generate_data(root, Positions)
generate_geometry_structure(root)
generate_step_structure(root)
ts = range(0,0.5,100)
for (iT, t) in enumerate(ts)
append_data(root, t, Positions)
end
end
f = h5open(“test.vtkhdf”, “w”)
root = HDF5.create_group(f, “VTKHDF”)
generate_data(root, Positions)
display(f)
close(f)
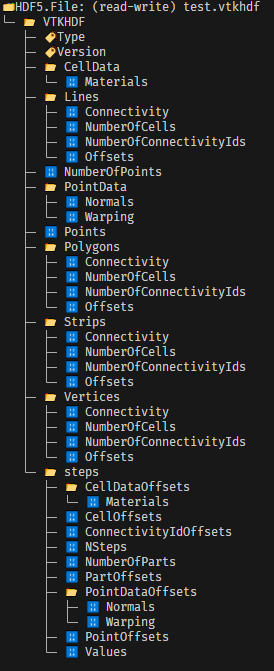
This will produce a few points in space, which will displace 100 time steps with a rand factor multiplied, just to simulate some motion. This example excels in showing:
I will have to be honest and say that this took way too long to figure out. The guide should really have been a simple step by step, showing how to add a few points. This took me ~3 months of thinking until I had understood enough with the format, that I could sit down with a hdfviewer software and continously compare the Python output and Julia one until it made sense / matches.
Why did I care so much about having it? On Windows, opening and closing a lot of vtkhdf files, is so extremely slow. For example for my simulation software, it would take 29 s (!) to close 1600 files. By saving data into one, it would take only ~0.5s!
I understand that funding is important for VTK to succeed etc., but please do not forget that all of us in open source, personal projects or even non directly funded commercial endevaours, by using the formats you provide, provide a legitimacy to the software format - and VTKHDF has been one of the harder file formats to crack, so I hope VTK does a bit more to promote it, because it is actually really great - just way way way too difficult, for someone who is not a VTK expert.