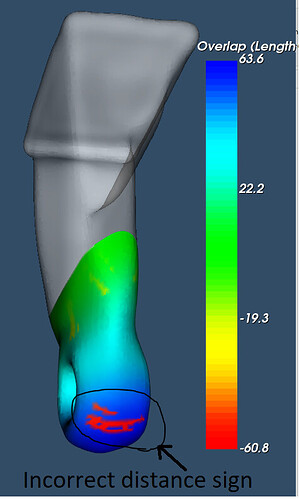
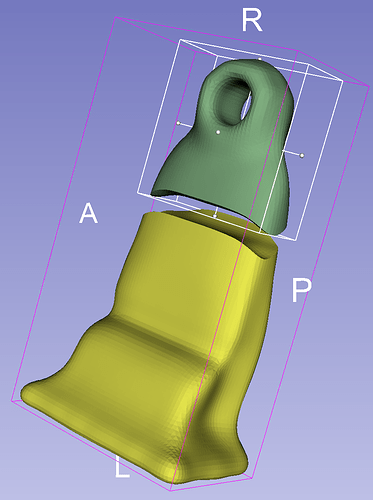
For the attached stl files, vtkDistancePolyDataFilter with signed distance ON produces incorrect signs on certain points. I could see the magnitude of the distance is correct, only the sign is flipped wrong.
Slightly translating the two meshes (that originally touched each other at that matching cut surface) to make the geometry better visible:
This explains what the problem is. vtkDistancePolyDataFilter uses vtkImplicitPolyDataDistance internally, which computes distance sign as “the sign of the dot product between the angle-weighted pseudonormal at the nearest surface point and the vector x - p”. This is unstable if you give such an ill-posed problem (where the vector connecting the closest points is nearly parallel with the surface normal).
If you really absolutely need signed distance then you may try to use vtkImplicitPolyDataDistance and adjust tolerance, etc. You may also add more heuristics (e.g., sign computation becomes unstable when you get far from the other model, so force sign of points that are above a certain threshold). Or, you can also make the computation fully robust by computing distance map over an image (rasterize the model and compute signed distance on the image). However, I think the proper solution would be to reformulate the question to something more meaningful (for example, ask for absolute distance when you measure distance between two non-intersecting meshes).
Thanks for the reply!
The mesh I have will always intersect, how does asking absolute distance helps?
This sign computation also gets unstable on points that are not so far from other model.
The model I shared looks easy to force sign above certain threshold, there are models which blends together (like a woven rope).
Just curious, If this is unstable how does the Boolean operations succeed? (https://www.vtkjournal.org/browse/publication/797)
Unfortunately, Boolean operations in VTK do not work reliably - they fail too often for relatively simple, completely valid inputs.
@lassoan could you please explain your comment at bit further:
Or, you can also make the computation fully robust by computing distance map over an image (rasterize the model and compute signed distance on the image)
I’m working with 3D image data objects in vtk, and extracting surfaces from signed distance fields on the vtkImageData from input polydatas using flying edges from vtk. But my signed distance field comes from vtkImplicitPolyDataDistance. I’d like to make more robust my signed distance computation if I can - could you explain your comment above a bit more so I could try to implement it using existing vtk filters? If possible if you can share code that you would use instead of vtkImplicitPolyDataDistance when operating on a vtkImageData, to get the same end result of a signed distance, that would be fantastic.
If you operate on images then you can compute distance map directly from the image. It seems that VTK’s vtkImageEuclideanDistance can only compute squared distance. If you need signed distance then you can use other libraries, such as ITK, which has several signed distance map computation methods, for example signed Maurer distance.
@lassoan thank you for your reply. I have not used ITK before, although have heard of ITK. I’m extracting the surface using vtk flying edges, is that available from ITK? If not, can ITK write a “.vti” vtkImageData file?
To get me off the ground and running, what would be the equivalent code in ITK to produce the signed distance? How do I convert the following to ITK:
imageData = vtkImageData()
imageData.SetDimensions(size)
imageData.SetSpacing(spacing)
imageData.SetOrigin(origin)
implicitDistance = vtkImplicitPolyDataDistance()
implicitDistance.SetInput(reader.GetOutput())
noClosestPoint = [1.0e10, 1.0e10, 1.0e10]
# set tolerance
implicitDistance.SetNoClosestPoint(noClosestPoint)
signed_distance = vtkDoubleArray()
signed_distance.SetName("signed_distance")
signed_distance.SetNumberOfComponents(1)
signed_distance.SetNumberOfTuples(imageData.GetNumberOfPoints())
imageData.GetPointData().AddArray(signed_distance)
iter = vtkImagePointIterator(imageData)
while not iter.IsAtEnd():
x, y, z = iter.GetPosition()
distance = implicitDistance.EvaluateFunction([x, y, z])
signed_distance.SetValue(iter.GetId(), distance)
iter.Next()
Many thanks
You can use files to pass data between ITK and VTK, but you need to choose a format that both ITK and VTK can read/write. vti is not among them, but there are several others that you can use, for example .mha and .nrrd. Running an ITK filter in Python should be straightforward, you can find lots of examples online.
If you work with images (the shapes are generated by segmenting an image) then you can avoid conversion from mesh to distance map and you can compute signed distance map directly from the image (using ITK filters). If your inputs are surface meshes (e.g., from a surface scanner) then you may have to use angle-weighted pseudonormals (such as vtkImplicitPolyDataDistance) or rasterization (such as vtkPolyDataToImageStencil) to create a distance map - which might fail if the mesh quality is not optimal.